Molecular Biophysics
Bojan Zagrovic
Group Leader
bojan.zagrovic@univie.ac.at
Phone: +43-1-4277-52271
Campus Vienna Biocenter 5, 1030 Vienna | Room: 1.601

Research
Research
The function of biomolecules arises from the interplay between their structure, dynamics and interactions with the environment. We explore this fundamental principle through the use of computational and theoretical methods, including molecular dynamics (MD) simulations and structural bioinformatics techniques, in close collaboration with experimentalists. The work in our group is organized around three principal directions.
First, we are interested in the role of dynamics and conformational entropy in non-covalent biomolecular interactions. We develop new methods for calculating conformational entropy of biomolecules from computer simulations and for measuring it experimentally. In addition to function, dynamics also affects the very process of biomolecular structure determination. We use MD simulations to help interpret time- and ensemble-averaged X-ray and NMR experiments and analyze the impact of conformational averaging on the derived structures.
Second, all biomolecular processes occur in crowded, dynamic, constantly changing environments. We study how crowding affects biomolecular interactions and other basic processes such as protein folding or post-translational modifications of proteins. In particular, we are interested in studying how crowding affects the co-localization of binding partners and employ MD and Brownian dynamics simulations and structural bioinformatics methods to address this question.
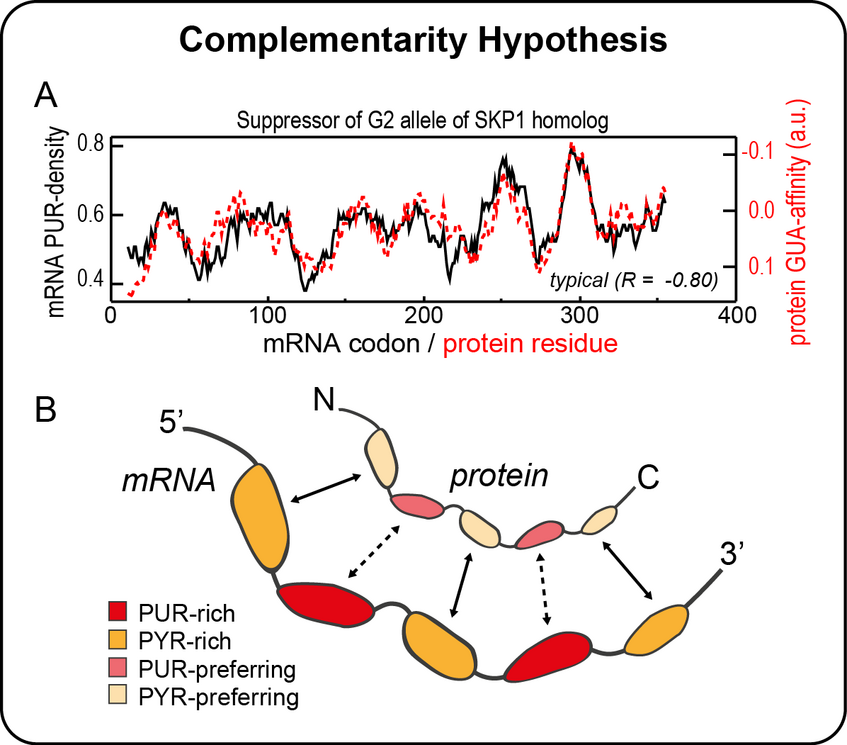
Finally, we have recently discovered a remarkably robust correspondence between the nucleobase density profiles of mRNAs and the nucleobase affinity profiles of their cognate protein sequences. For example, the purine-density profile of a typical mRNA coding sequence in H. sapiens matches the guanine-affinity profile of its cognate protein with an absolute value of the Pearson correlation of 0.8 on average. We believe this finding supports and extends the stereochemical hypothesis concerning the origin of the genetic code and suggests that cognate mRNAs and proteins may be physico-chemically complementary to each other and bind, especially if unstructured. Moreover, this findings suggests a novel principle of RNA-protein interactions beyond the cognate context, with potential implications in different areas of RNA-protein biology. We use different biophysical methods including MD simulations, structural bioinformatics techniques, free energy calculations and in vitro experiments to further explore this hypothesis.
Publications
Polyansky AA & Zagrovic B (2013). Evidence of direct complementary interactions between messenger RNAs and their cognate proteins. NUCLEIC ACIDS RES;41(18):8434-8443. PMID: 23868089
de Ruiter, Anita; Zagrovic, Bojan (2015). Absolute binding-free energies between standard RNA/DNA nucleobases and amino-acid sidechain analogs in different environments. NUCLEIC ACIDS RES;43(2):708-18. PMID: 25550435
Fleck, Markus; Polyansky, Anton A; Zagrovic, Bojan (2016). PARENT: A Parallel Software Suite for the Calculation of Configurational Entropy in Biomolecular Systems. J CHEM THEORY COMPUT;12(4):2055-65. PMID: 26989950
more publications of the group Zagrovic
Collaborations & Funding

Doctoral Program "Integrative Structural Biology"
2016-2019: The Group Zagrovic participates in the special Doctoral Program "Integrative Structural Biology" reviewed and funded by the Austrian Science Fund FWF.

ERC Starting Grant 2011
Awardee of a "Starting Independent Researcher Grant" from the European Research Council ERC

START Prize 2010, Austrian Science Fund (FWF)
Project title: "Specific and global aspects of protein interactions"
