Structural Biology of the Cytoskeleton
Kristina Djinovic-Carugo
Group Leader
kristina.djinovic@univie.ac.at
Phone: +43-1-4277-52203
Campus Vienna Biocenter 5 (VBC 5), 1030 Vienna | Room: 1.606

Research
Research
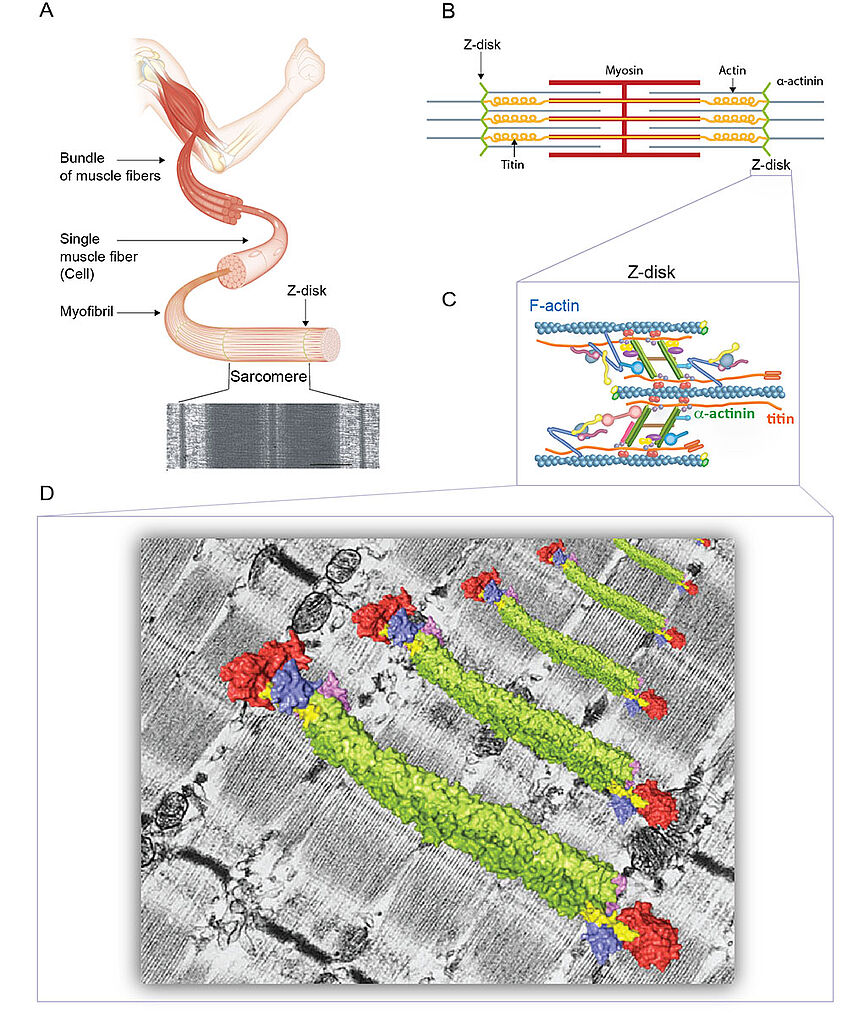
Animal movement is mediated by striated muscles, with the sarcomere being the basal contractile unit within single muscle fibers. Sarcomeric actin and myosin form cross-linked and interdigitated filament bundles whose sliding motion generates force. Antiparallel actin filaments from adjacent sacromers are anchored at the Z-disk, which plays a central role as the site organizing the molecular machinery that is required for muscle contraction. The Z-disk, a stable and highly ordered protein network is both the mechanical hub of the system and a signaling platform, and is regarded as one of the most complex assemblies known to biology (Figure 1A-C).
Much is known about the pairwise interactions of the major Z-disk components, less about strcutures of Z-disk components, but due to its complexity little is known about their assembly into higher-order structures and ultimately the Z-disk itself. The major questions we are addressing are: What is the stoichiometry of the components and the assembly hierarchy of the Z-disk? What is the molecular architecture of both pre- and myofibrillar assemblies? Our long-term goal is to reconstitute a functional Z-disk from individual components, which is the best way to truly understand their disparate functional roles and molecular mechanisms. We recently determined the crystal structure of the major component of the Z-diak - human muscle α-actinin-2, providing insight into the mechanism that promotes the molecular assembly of the Z-disk (Cell, 2014) (Figure 1D).
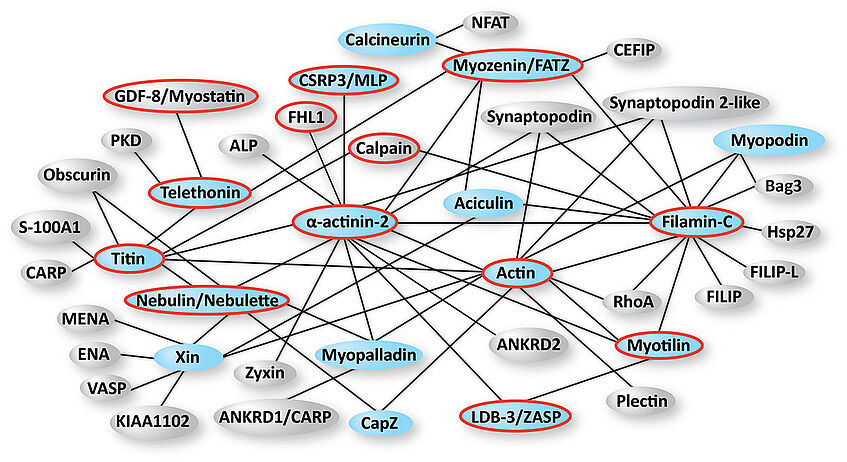
Apart from its major component α-actinin-2, which accounts for ∼20% of the Z-disk mass, the current inventory of proteins in mature Z-disks includes over 40 proteins that form a highly stable and highly ordered unit that can support contractile forces of the muscle. The production of 16 Z-disk proteins is already well established in our laboratory (Figure 2).
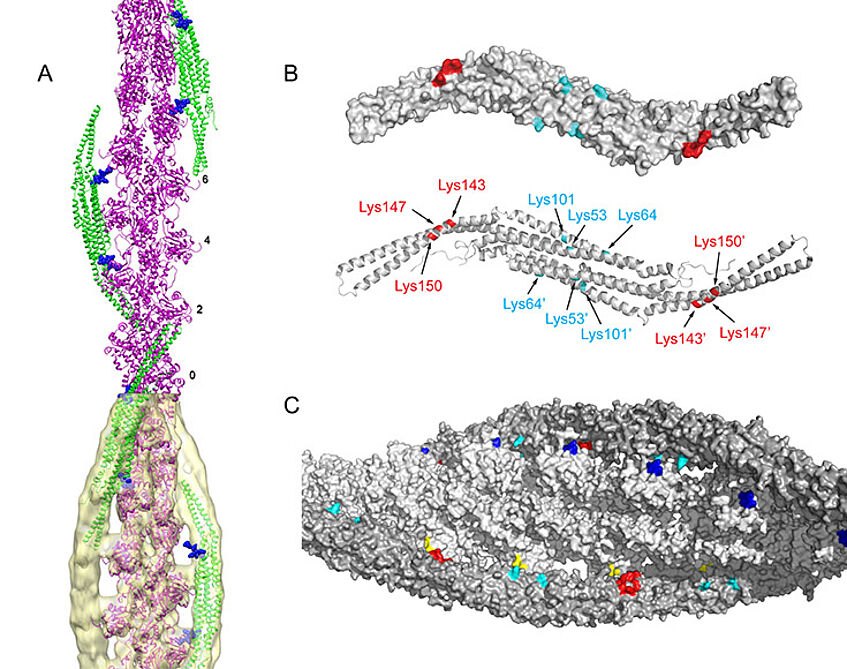
We are proceeding with generation of binary and higher complexes and their biochemical, biophysical and structural characterisation combining high resolution studies (X-ray diffraction, NMR) with lower resolution approaches that can either yield molecular envelopes (SAXS, SANS, EM) or specific distance information (chemical cross-linking coupled to mass-spectrometry, NMR) (Figure 3).
These activities are complemented by the development of bioinformatics tools for results and fine tuning of the protein constructs to be structurally analyzed. New bioinformatics strategies are being designed to extend our prediction capabilities.
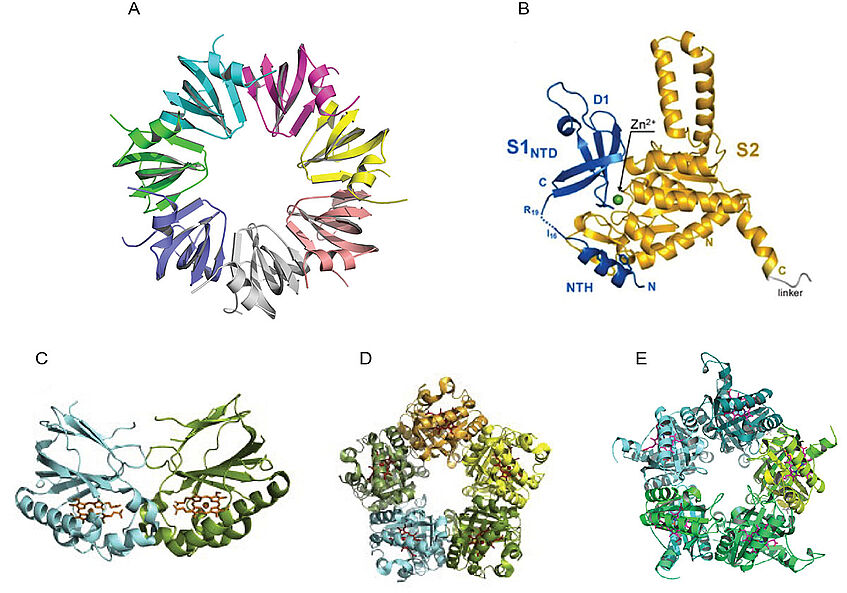
With other groups and faculties at MFPL we are working on structural studies of the RNA chaperone Hfq and its interactions with RNA (Bläsi), trypasonomal cytoskeletal protein MORN1 (Warren), ribosomal proteins S1-S2 (Moll) (Figure 4A,B), poly(ADP-ribose) glycohydrolase (PARG) (Slade) and vaccinia virus protein A46 Skern) and tristetraproline (TTP) (Kovarik).
In collaboration with M. Wagner (Faculty for Life Sciences, Univ. Vienna) and C. Obinger (University of Natural Resources and Life Sciences, Vienna) we are studying the family of bacterial chlorite dismutases (Figure 4C-D). With C. Obinger we furthermore collaborate structural studies of engeneered antibody Fc fragmets and their complexes with selected antigens.
In order to overcome the major bottlenecks in structural and functional studies of proteins, which are availability of milligram amounts of active, chemically and conformationally pure protein and crystallization, an FFG funded Laura Bassi Centre for Optimized Structural Studies (COSS) was established as a joint venture of IMP, Intercell and UNIVIE-MFPL with the goal to set-up an efficient platform to combine the recent advances in automated expression screening of protein targets employing nested constructs design and cell-free protein expression systems followed by biophysical characterization to find conditions best suited for structural and functional studies.
Publications
Grison, Marco; Merkel, Ulrich; Kostan, Julius; Djinovic-Carugo, Kristina; Rief, Matthias (2017). alpha-Actinin/titin interaction: A dynamic and mechanically stable cluster of bonds in the muscle Z-disk. P NATL ACAD SCI USA;114(5):1015-1020. PMID: 28096424
Ribeiro, Euripedes de Almeida; Pinotsis, Nikos; Ghisleni, Andrea; Salmazo, Anita; Konarev, Petr V; Kostan, Julius; Sjöblom, Björn; Schreiner, Claudia; Polyansky, Anton A; Gkougkoulia, Eirini A; Holt, Mark R; Aachmann, Finn L; Zagrović, Bojan; Bordignon, Enrica; Pirker, Katharina F; Svergun, Dmitri I; Gautel, Mathias; Djinović-Carugo, Kristina (2014). The Structure and Regulation of Human Muscle α-Actinin. CELL;159(6):1447-60. PMID: 25433700
Kostan J, Salzer U, Orlova A, Toeroe I, Hodnik V, Senju Y, Zou J, Schreiner C, Steiner J, Meriläinen J, Nikki M, Virtanen I, Carugo O, Rappsilber J, Lappalainen P, Lehto VP, Anderluh G, Egelman EH, Djinović-Carugo K (2014). Direct Interaction of Actin Filaments with F-BAR Protein Pacsin2. EMBO REP;15(11):1154-62. PMID: 25216944
more publications of the group Djinović
Collaborations & Funding

Christian Doppler Laboratory for knowledge-based structural biology and biotechnology
Project-leader: Kristina Djinović-Carugo and Robert Konrat
Business partner: Boehringer Ingelheim RCV GmbH & Co KG, Arsanis Biosciences GmbH and Biomin Holding GmbH
Duration: 01.02.2017 - 31.01.2024
CD Laboratory for knowledge-based structural biology and biotechnology
Christian Doppler Research Association
University of Vienna
Description: The scientists will primarily investigate protein structure, which is crucial for the development of new therapeutic methods for a great variety of diseases ranging from Alzheimer’s and infectious diseases to cancer. This renders it a most valuable tool in biomedical research. In order to gain new insights into protein structure, function and interactions that could potentially be translated into new therapies, the researchers will employ a combination of bioinformatics, protein production and high-end biophysical and structural biology techniques. The collected information will ultimately be combined in an information pipeline for both research and biotechnology.

Wellcome Trust Collaborative Award
2016-2020: An integrated approach to the muscle Z-disk: from atomic structure to human disease (with M. Gautel, P. Elliott, H. Watkins, S. Raunser and K Gehmlich)

Doctoral Program "Integrative Structural Biology"
2016-2019: The Group Djinovic participates in the special Doctoral Program "Integrative Structural Biology" reviewed and funded by the Austrian Science Fund FWF. Kristina Djinovic-Carugo is Vice Speaker of the program.
Doctoral Program "Structure and Interaction of Biological Macromolecules"
2009-2013: Doctoral Program "Structure and Interaction of Biological Macromolecules" reviewed and funded by the Austrian Science Fund FWF.

COSS - Center for Optimized Structural Studies
2014-2016: Laura-Bassi Center of Expertise "COSS - Center for Optimized Structural Studies" funded by the Austrian Research Promotion Agency FFG (Co-ordinator)
2010-2013: Laura-Bassi Center of Expertise "COSS - Center for Optimized Structural Studies" funded by the Austrian Research Promotion Agency FFG (Co-ordinator)

Bioinformatics Integration Network III
2009-2013: Research Network funded by the GEN-AU Genome Research Program of the Federal Ministry of Science and Researc

DFG/FWF Research Unit
2014-2017: "Structure and regulation of the Myofibrillar Z-disc Interactome" funded by the German Research Foundation and the Austrian Science Fund
2010-2014: Research Unit 'Alpha Actinin, Filamin C and their Complexes with Binding Partners' funded by the German Research Foundation and the Austrian Science Fund

Marie Curie Initial Training Network
2009-2014: FP7 Marie-Curie Action ITN Muscle Z-disk Protein Complexes: from atomic structure to physiological function (MUZIC)

FWF Stand Alone Projects
2014-2017: MORN “Structure and Function of the Trypanosoma brucei bilobe”
2010-2013: MORN repeat proteins and the Trypanosoma brucei bilobe (with G. Warren)
2010-2013: Structural Aspects of RAF-1:Rok-alpha Interaction (with M. Baccarini)

Doctoral Program "Signaling Mechanisms in Cellular Homeostasis"
The Group Djinović is an associated member of the special Doctoral Program "Signaling Mechanisms in Cellular Homeostasis" reviewed and funded by the Austrian Research Fund FWF.
